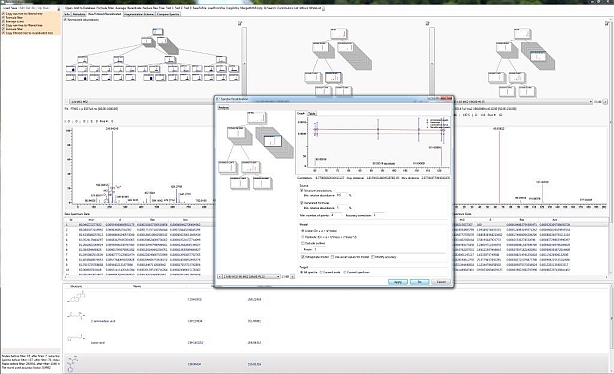
mzCloud™ is an extensively curated database of high-resolution tandem mass spectra that are arranged into spectral trees. MS/MS and multi-stage MSn spectra were acquired at various collision energies, precursor m/z, and isolation widths using Collision-induced dissociation (CID) and Higher-energy collisional dissociation (HCD). Each raw mass spectrum was filtered and recalibrated resulting in recalibrated spectral trees that are fully searchable. Besides the experimental and processed data, each database record contains the compound name with synonyms, the chemical structure, computationally and manually annotated fragments (peaks), identified adducts and multiply charged ions, molecular formulas, predicted precursor structures, detailed experimental information, peak accuracies, mass resolution, InChi, InChiKey, and other identifiers. mzCloud is a fully searchable library that allows spectra searches, tree searches, structure and substructure searches, and more. To preview all mass spectra or conduct searches, please go to Database.
- Spectral trees – intuitively organized multi-stage tandem mass spectra
- Identification of compounds even if they are not represented in the library
- Structurally annotated spectral peaks using non-trivial in silicio prediction methods
- Smart substructure search algorithms capable of extracting spectra with common structural scaffolds
- Fragment structure support throughout the database
- Extensive experimental information and identifier metadata data is included
- Rigorously curated data
Spectral trees – intuitively organized multi-stage tandem mass spectra
Soft ionization and CID techniques can generate spectra whose appearance depends on the experimental conditions and sample preparation. To manage and search diverse product spectra with an identical precursor ion for a single chemical entity, mzCloud assembles data into spectral trees Each node collects all spectrum of an identical precursor m/z value acquired at different conditions.
If a node contains more than two parallel spectra, the application automatically calculates the average and composite spectra.
The spectral node strategy strengthens the robustness of all the mathematical processing methods and, compared to simple spectra averaging, does not distort the highly nonlinear peak ratio progress.
You can easily access node spectra from a library by clicking the edge of the spectra, or by browsing in the box displayed below the tree.
Identification of compounds even if they are not represented in the library
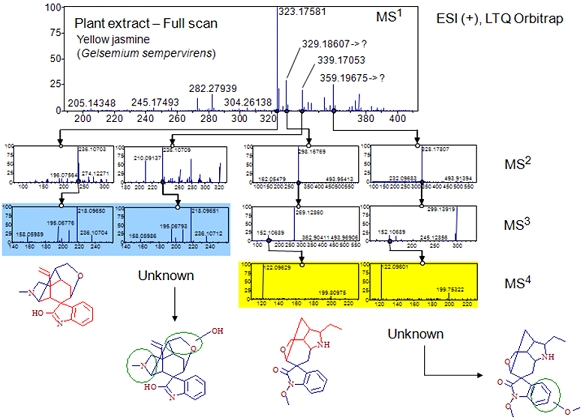
mzCloud is the first public spectral database that offers the Precursor Ion Fingerprinting (PIF) technique. This innovative approach identifies substructural information through the comparison of product ion spectra of structurally related compounds. Structural information is derived by utilizing previously characterized ion structures stored in mzCloud and matching them with unknown product ion spectra. PIF is a very powerful technique that heavily depends upon spectra of precursor ions of various chemical classes acquired under various experimental conditions. The more ion fingerprints present in the library, the more substructures can be identified from unknown spectra.
 If you would like to learn more about PIF, please refer to the following resources:
If you would like to learn more about PIF, please refer to the following resources:
- Determination of Molecular Structures Using Tandem Mass Spectrometry. U.S. 7197402 B2, 2007
- Determination of Ion Structures Using PIF: J Am Soc Mass Spectrom 2009, 20, 370–376
- PIF: Precursor Ion Fingerprinting – Searching for a Structurally Diagnostic Fragment Using Combined Targeted and Data Dependent MSn
To empower this approach, it is important to acquire the substructural fingerprints represented as spectral treesthat cover a significant portion of biologically, environmentally or toxicologically relevant structural space, contributors from various metabolomics, natural compound, forensic or industrial fields are welcome others to contribute to the mzCloud database.
Structurally annotated spectral peaks using non-trivial in silicio prediction methods
Annotations not only allow a better understanding of the fragmentation processes which led to a particular peak, but they also provide a useful tool for the identification of unknowns.
To determine the correct identity of the fragment structures needed for the substructural characterization of precursor ions, all fragments in the mzCloud database are predicted using advanced heuristic and a broad range of quantum chemical methods:
- advanced prediction of fragments using Mass Frontier, widely recognized as the industry’s leading software, by applying general fragmentation rules and more than a hundred thousand mechanisms which have been published in peer-reviewed journals
- manual assignment and evaluation of fragments by an experienced mass spectrometrist
- refinement of predicted fragments using complex reaction step calculations by applying ab initio quantum chemical methods. This approach enables the assignment of realistic fragment structures to an observed peak by assigning relative probabilities to different pathways. On top of this, approximate intensities are estimated and compared to the spectral peaks, and serve as additional confirmation/rejection criteria
For the assignment of fragment structures to a precursor ion in the process of creating structurally characterized product ion spectra, it is extremely beneficial to have high-resolution spectra since the accurate m/z values of precursor and product ions greatly reduce the number of possible molecular formulas for fragment structures. Also, the determination of the structural arrangement for the elucidated molecule benefits from exact mass measurements by constraining the elemental composition of the elucidated molecule and consistently validating the calculated mass of recognized fragment structures and accurate m/z values of precursor and product ions.
Smart substructure search algorithms capable of extracting spectra with common structural scaffolds
Structure and substructure searches are useful features for retrieving library entries with specific structural moieties. mzCloud allows tailored substructural searches by applying various search rules. Alternatively, substructure searches can find groups of potentially fragmetationally related compounds. Because fragmentation mechanisms on rings significantly differ from acyclic moieties, searching substructures that exactly match the ring membership and have the best overlap with library structures can further refine the search results. In doing so, the Substructure Match Ring Bonds options available in mzCloud yield structures, which are mass spectrometrically reasonable.
Fragment structure support throughout the database
In addition to neutral molecules, fragment structures, including protonated, deprotonated and adduct molecules, are also supported throughout the mzCloud databases. The integrated structure editor allows fragment structures to be drawn and searched in libraries. The peaks of reference spectra are also annotated with fragments. Isotope-labeled compounds are supported as well.
Extensive experimental information and identifier metadata data is included
As with any professional reference database, mzCloud keeps and displays extensive supplementary information and data sources along with experimental parameters and a collection of compound identifiers.
Rigorously curated data
Only high-quality data can lead to reliable and accurate results. Therefore, we take the data processing of raw data very seriously. All spectra, structures and metadata are rigorously manually and electronically evaluated for errors, checked for consistency and correctness, filtered, recalibrated and compound identifiers are compared with external databases. Full-time curators with a mass spectrometric background process every single spectrum manually. Dedicated software with a number of novel algorithms is used for transforming raw data into high quality spectral trees.